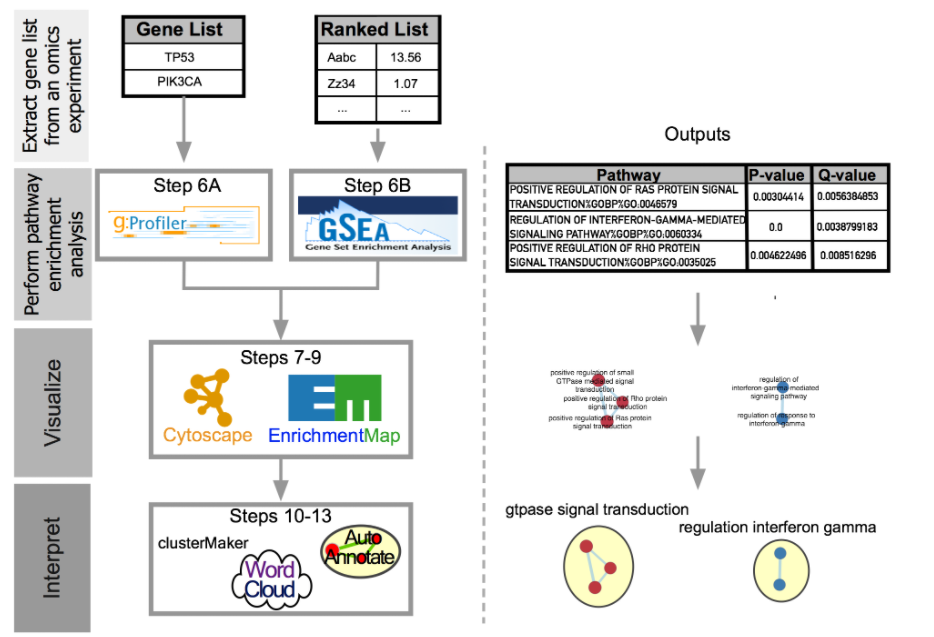
Enrichment Map是一个用于功能富集可视化的Cytoscape插件,用于帮助可视化和分析由Gene Set Enrichment Analysis(GSEA)、BiNGO或David等程序生成的功能富集结果。
下面是Enrichment Map的一个标准的流程:
官方学习素材:
- Download TCGA data – R notebook shows you how to download legacy microarray and rnaseq ovarian cancer data. The notebook can be modified to download any data from GDC
- Supplemental Protocol 1 – convert raw RNASeq expression data to a ranked list
- Supplemental Protocol 2 – convert RMA normalized microarray expression data to a ranked list
- Supplemental Protocol 3 – perform Pathway Enrichment Analysis in R using ROAST and Camera
- Supplemental Protocol 4 – perform phenotype randomizations using edgeR with GSEA.
- Supplemental Protocol 5 – Creates a Multi dataset Enrichment Map (using GSEA results from multiple rank files) and followed by Theme calculation and summary.
- Protocol 2 – run GSEA on ranked list and automatically create an Enrichment Map from the results.
官方示例文件:
- Supplmentary table 1 – Cancer drivers. List of genes used as g:Profiler input.
- Supplmentary table 2 – GSEA Rank file. List of genes and their assoicated ranks used as input for GSEA.
- Supplmentary table 3 – GSEA GMT file. Pathway defintion file used for the protocol as GSEA input. Updated gmt files can be found on the baderlab download site here
- Supplmentary table 4 – Example g:Profiler results file.
- Supplmentary table 5 – Example g:Profiler gmt file. If using example g:Profiler results file instead of regenerating the results make sure to use this gmt file. If the g:Profiler results file has been regenerated then make sure to download an updated gmt file that corresponds to the results file as specified in the protocol.
- Supplmentary table 6 – Example expression file (corresponding to example rank file, supplementary 2) used when creating an Enrichment Map from GSEA results.
- Supplmentary table 7 – Example class file (corresponding to example expression file, supplementary 6) used when creating an Enrichment Map from GSEA results.
- Supplmentary table 8 – Example GSEA results file 1.
- Supplmentary table 9 – Example GSEA results file 2.
- Supplmentary table 10 – Example RANSeq expression input file used in Supplementary Protocol 1. File used to generate rank file, supplementary table 2, used in the main protocol.
- Supplmentary table 11 – Example sample definition input file used in Supplementary Protocol 1. File used to generate rank file, supplementary table 2, used in the main protocol.
- Supplmentary table 12 – Example Microarray expression input file used in Supplementary Protocol 2.
- Supplmentary table 13 – Example sample definition input file used in Supplementary Protocol 2.
利用上面的数据,可以根据教程:https://enrichmentmap.readthedocs.io/en/latest/来进行学习。
参考资料:
1.http://baderlab.org/GeneSets
2.https://baderlab.github.io/Cytoscape_workflows/EnrichmentMapPipeline/index.html
 浙公网安备 33010802011761号
浙公网安备 33010802011761号