前面文章介绍了R GSEA(Gene Set Enrichment Analysis)快速入门,当我们用GSEA软件做数据分析的时候,对其文件格式却知之甚少,本文根据官方的介绍做了汇总整理。
软件下载:
| GSEA v4.1.0 Mac App | Download and unzip the Mac App Archive then double-click the GSEA application to run it. You can move the app to the Applications folder or anywhere else. | download GSEA_4.1.0.app.zip |
|---|---|---|
| GSEA v4.1.0 for Windows | Download and run the installer. A GSEA shortcut will be created on the Desktop; double-click it to run the application. 64-bit Windows is required | download GSEA_Win_4.1.0-installer.exe |
| GSEA v4.1.0 for Linux | Download and unzip the Archive. See the included readme.txt for further instructions. 64-bit Linux is required | download GSEA_Linux_4.1.0.zip |
| GSEA v4.1.0 for the command line (all platforms) | Download and unzip the Archive. See the included readme.txt for further instructions. Requires separate Java 11 installation. | download GSEA_4.1.0.zip |
| GSEA v4.1.0 Java Web Start (all platforms) | Launches the GSEA desktop application from the web. Requires separate Java 8 installation. Please use a configuration smaller than your computer’s total memory. This option will be removed in a future release. | Launch with 1GB (for 32 or 64-bit Java) 2GB (for 64-bit Java only) 4GB (for 64-bit Java only) 8GB (for 64-bit Java only) |
主要涉及5类文件格式:
1)表达矩阵文件
GCT文件(比较常用): Gene Cluster Text file format (*.gct)
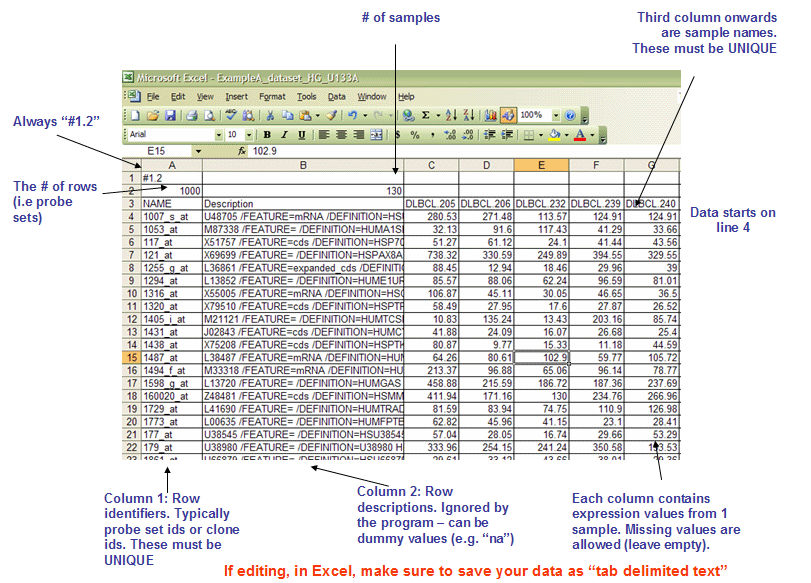
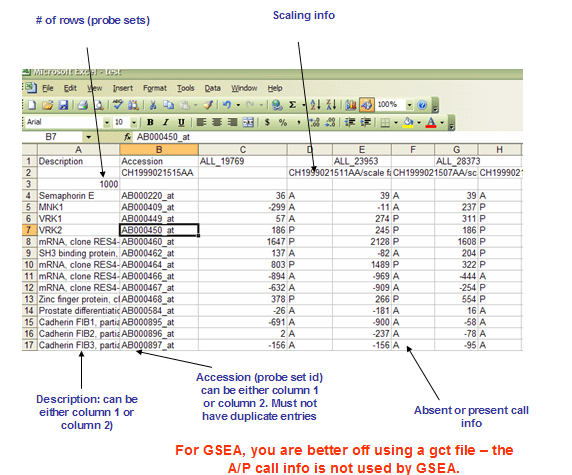
RES文件: ExpRESsion (with P and A calls) file format (*.res)
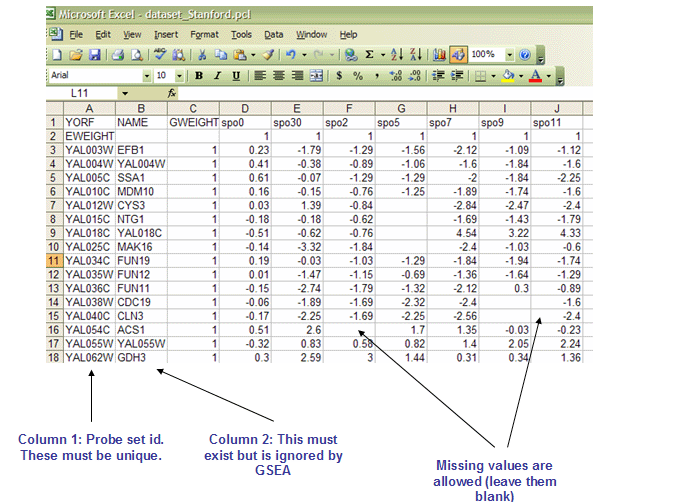
PCL文件: Stanford cDNA file format (*.pcl)
TXT文件( 推荐 ): Text file format for expression dataset (*.txt)
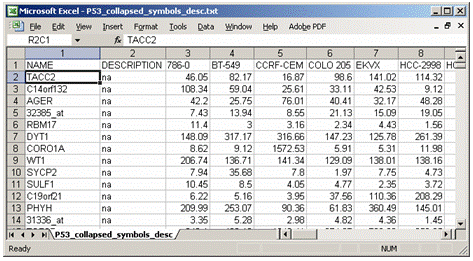
注意:第一列和第二列表头必须为NAME、 Description
2)表型文件
CLS文件: Categorical (e.g tumor vs normal) class file format (*.cls)

3)基因集数据库文件
GMX文件: Gene MatriX file format (*.gmx)
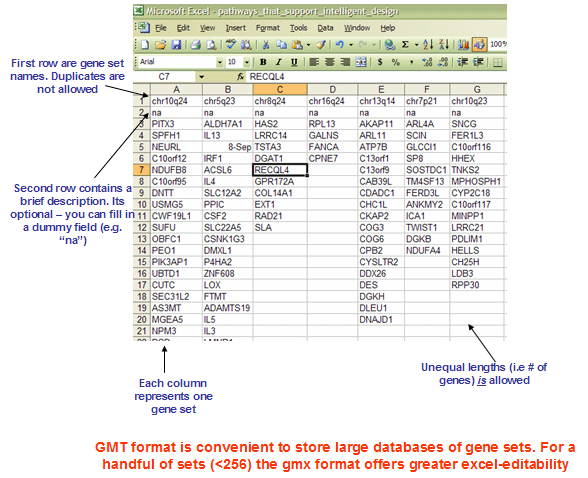
GMT文件: Gene Matrix Transposed file format (*.gmt)

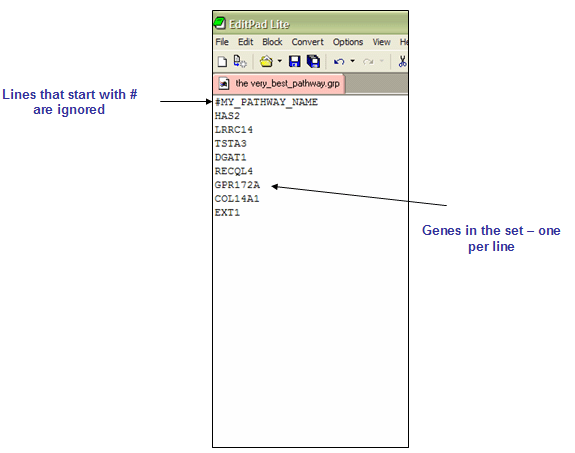
GRP文件: Gene set file format (*.grp)
XML文件: Molecular signature database file format (msigdb_*.xml)

4)芯片注释文件(现在芯片使用相对较少)
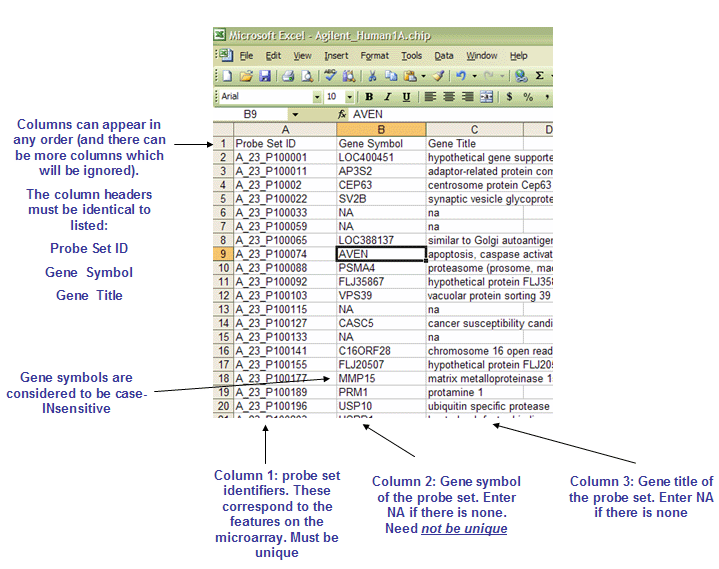
CHIP文件: Chip file format (*.chip)
5)基因的排名文件(输出结果会包含)
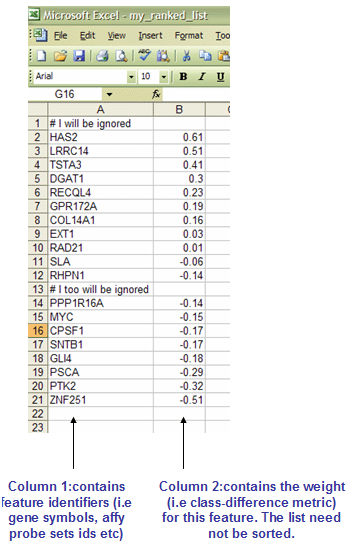
RNK文件: Ranked list file format (*.rnk)
当你了解了GSEA涉及的各种文件后,使用GSEA软件将会更加容易。
参考资料:
1.http://software.broadinstitute.org/cancer/software/gsea/wiki/index.php/Data_formats
2.http://www.broadinstitute.org/gsea/
 浙公网安备 33010802011761号
浙公网安备 33010802011761号