这个问题是源于在批量处理RNA-seq数据时使用hisat2时捕获输出的统计信息。
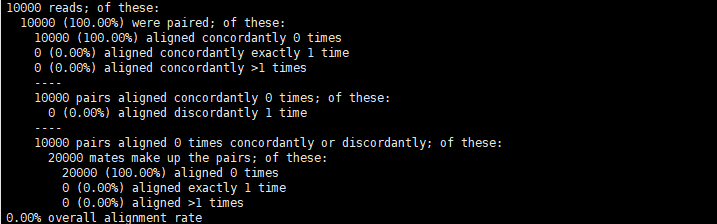
一般来说,这段信息直接在标准输出下,既不是返回值也不是退出的文本信息。
linux中有三种标准输入输出,分别是STDIN,STDOUT,STDERR,对应的数字是0,1,2。
STDIN是标准输入,默认从键盘读取信息;
STDOUT是标准输出,默认将输出结果输出至终端;
STDERR是标准错误,默认将输出结果输出至终端。
官方说明:
When HISAT2 finishes running, it prints messages summarizing what happened. These messages are printed to the “standard error” (“stderr”) filehandle. For datasets consisting of unpaired reads, the summary might look like this……
我们先找到Hisat2中报告生成的代码片段
https://github.com/DaehwanKimLab/hisat2/blob/master/aln_sink.h
部分源码如下:
if(newSummary) {
out << "HISAT2 summary stats:" << endl;
if(totpair > 0) {
uint64_t ncondiscord_0 = met.nconcord_0 - met.ndiscord;
out << "\tTotal pairs: " << totpair << endl;
out << "\t\tAligned concordantly or discordantly 0 time: " << ncondiscord_0 << " ("; printPct(out, ncondiscord_0, met.npaired); out << ")" << endl;
out << "\t\tAligned concordantly 1 time: " << met.nconcord_uni1 << " ("; printPct(out, met.nconcord_uni1, met.npaired); out << ")" << endl;
out << "\t\tAligned concordantly >1 times: " << met.nconcord_uni2 << " ("; printPct(out, met.nconcord_uni2, met.npaired); out << ")" << endl;
out << "\t\tAligned discordantly 1 time: " << met.ndiscord << " ("; printPct(out, met.ndiscord, met.npaired); out << ")" << endl;
out << "\tTotal unpaired reads: " << ncondiscord_0 * 2 << endl;
out << "\t\tAligned 0 time: " << met.nunp_0_0 << " ("; printPct(out, met.nunp_0_0, ncondiscord_0 * 2); out << ")" << endl;
out << "\t\tAligned 1 time: " << met.nunp_0_uni1 << " ("; printPct(out, met.nunp_0_uni1, ncondiscord_0 * 2); out << ")" << endl;
out << "\t\tAligned >1 times: " << met.nunp_0_uni2 << " ("; printPct(out, met.nunp_0_uni2, ncondiscord_0 * 2); out << ")" << endl;
} else {
out << "\tTotal reads: " << totread << endl;
out << "\t\tAligned 0 time: " << met.nunp_0 << " ("; printPct(out, met.nunp_0, met.nunpaired); out << ")" << endl;
out << "\t\tAligned 1 time: " << met.nunp_uni1 << " ("; printPct(out, met.nunp_uni1, met.nunpaired); out << ")" << endl;
out << "\t\tAligned >1 times: " << met.nunp_uni2 << " ("; printPct(out, met.nunp_uni2, met.nunpaired); out << ")" << endl;
}
out << "\tOverall alignment rate: "; printPct(out, tot_al, tot_al_cand); out << endl;然后我们找到对应的调用文件:
https://github.com/DaehwanKimLab/hisat2/blob/master/aln_sink.cpp
部分代码:
assert_neq(ReportingState::EXIT_SHORT_CIRCUIT_TRUMPED, exitUnpair1_);
assert_neq(ReportingState::EXIT_SHORT_CIRCUIT_TRUMPED, exitUnpair2_);
if((paired_ && !p_.mixed) || nunpair1_ + nunpair2_ == 0) {
// Unpaired alignments either not reportable or non-existant
return;
}
// Do we have 1 or more alignments for mate #1 to report?
if(exitUnpair1_ == ReportingState::EXIT_SHORT_CIRCUIT_k) {
// k at random
assert_geq(nunpair1_, (uint64_t)p_.khits);
nunpair1Aln = p_.khits;
} else if(exitUnpair1_ == ReportingState::EXIT_SHORT_CIRCUIT_M) {
assert(p_.msample);
assert_gt(nunpair1_, 0);
unpair1Max = true; // repetitive alignments for mate #1
nunpair1Aln = 1; // 1 at random
} else if(exitUnpair1_ == ReportingState::EXIT_WITH_ALIGNMENTS) {
assert_gt(nunpair1_, 0);
// <= k at random
nunpair1Aln = min<uint64_t>(nunpair1_, (uint64_t)p_.khits);
}简单来看,可以看出这里采用了assert(assert的表达式(等于0或者为false时,assert会向stderr输出一些错误信息)所以常规我们使用>>log.txt(重定向标准输出)是无法将信息定向到文件中去的。
hisat2的这段输出信息相当于输出到 STDERR ,所以我们要将这段信息定义到文本中,采用如下方式:
hisat2 -p 4 -x ./grch38_tran/genome_tran -1 ./reads_1.fq -2 ./reads_2.fq -S ./test.sam >>log.txt 2>&1解决问题!
参考资料:
1.https://ccb.jhu.edu/software/hisat2/manual.shtml#reporting
 浙公网安备 33010802011761号
浙公网安备 33010802011761号