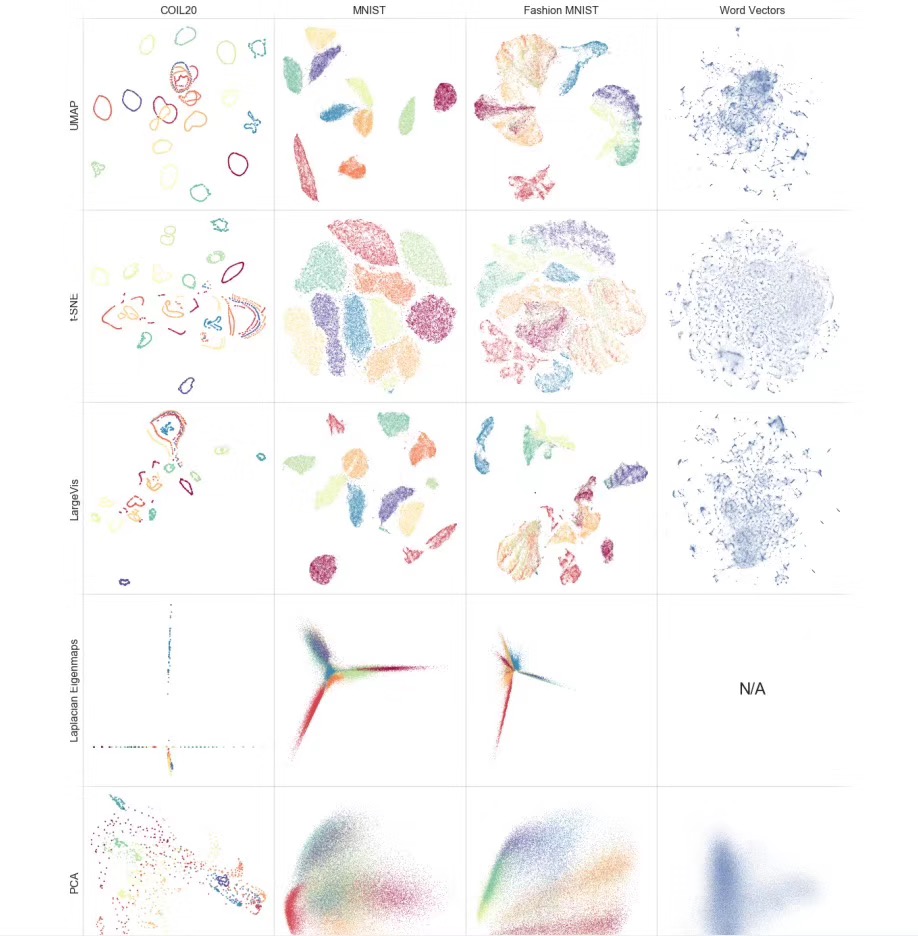
说到UMAP,不得不提一下t-SNE。UMAP是类似于t-SNE的一种数据降维算法
UMAP优点,简要来说有以下两点:
- 非线性降维,速度快(比t-SNE快很多,尤其在高维变量)
- 支持更多的变量/更高维度
下面我们也介绍下UMAP的R包使用方法:
step1.安装UMAP包
install.packages("uwot")step2.一个示例
library(uwot)
# See function man page for help
?umap
# Non-numeric columns are ignored, so in a lot of cases you can pass a data
# frame directly to umap
iris_umap <- umap(iris, n_neighbors = 50, learning_rate = 0.5, init = "random")
# Load mnist from somewhere, e.g.
# devtools::install_github("jlmelville/snedata")
# mnist <- snedata::download_mnist()
mnist_umap <- umap(mnist, n_neighbors = 15, min_dist = 0.001, verbose = TRUE)
# For high dimensional datasets (> 100-1000 columns) using PCA to reduce
# dimensionality is highly recommended to avoid the nearest neighbor search
# taking a long time. Keeping only 50 dimensions can speed up calculations
# without affecting the visualization much
mnist_umap <- umap(mnist, pca = 50)
# Use a specific number of threads
mnist_umap <- umap(mnist, n_neighbors = 15, min_dist = 0.001, verbose = TRUE, n_threads = 8)
# Use a different metric
mnist_umap_cosine <- umap(mnist, n_neighbors = 15, metric = "cosine", min_dist = 0.001, verbose = TRUE, n_threads = 8)
# If you are only interested in visualization, `fast_sgd = TRUE` gives a much faster optimization
mnist_umap_fast_sgd <- umap(mnist, n_neighbors = 15, metric = "cosine", min_dist = 0.001, verbose = TRUE, fast_sgd = TRUE)
# Supervised dimension reduction
mnist_umap_s <- umap(mnist, n_neighbors = 15, min_dist = 0.001, verbose = TRUE, n_threads = 8,
y = mnist$Label, target_weight = 0.5)
# Add new points to an existing embedding
mnist_train <- head(mnist, 60000)
mnist_test <- tail(mnist, 10000)
# You must set ret_model = TRUE to return extra data we need
# coordinates are in mnist_train_umap$embedding
mnist_train_umap <- umap(mnist_train, verbose = TRUE, ret_model = TRUE)
mnist_test_umap <- umap_transform(mnist_test, mnist_train_umap, verbose = TRUE)
# Save the nearest neighbor data
mnist_nn <- umap(mnist, ret_nn = TRUE)
# coordinates are now in mnist_nn$embedding
# Re-use the nearest neighor data and save a lot of time
mnist_nn_spca <- umap(mnist, nn_method = mnist_nn$nn, init = spca)
# No problem to have ret_nn = TRUE and ret_model = TRUE at the same time
# Calculate Petal and Sepal neighbors separately (uses intersection of the resulting sets):
iris_umap <- umap(iris, metric = list("euclidean" = c("Sepal.Length", "Sepal.Width"),
"euclidean" = c("Petal.Length", "Petal.Width")))
# Can also use individual factor columns
iris_umap <- umap(iris, metric = list("euclidean" = c("Sepal.Length", "Sepal.Width"),
"euclidean" = c("Petal.Length", "Petal.Width"),
"categorical" = "Species"))step3.绘图
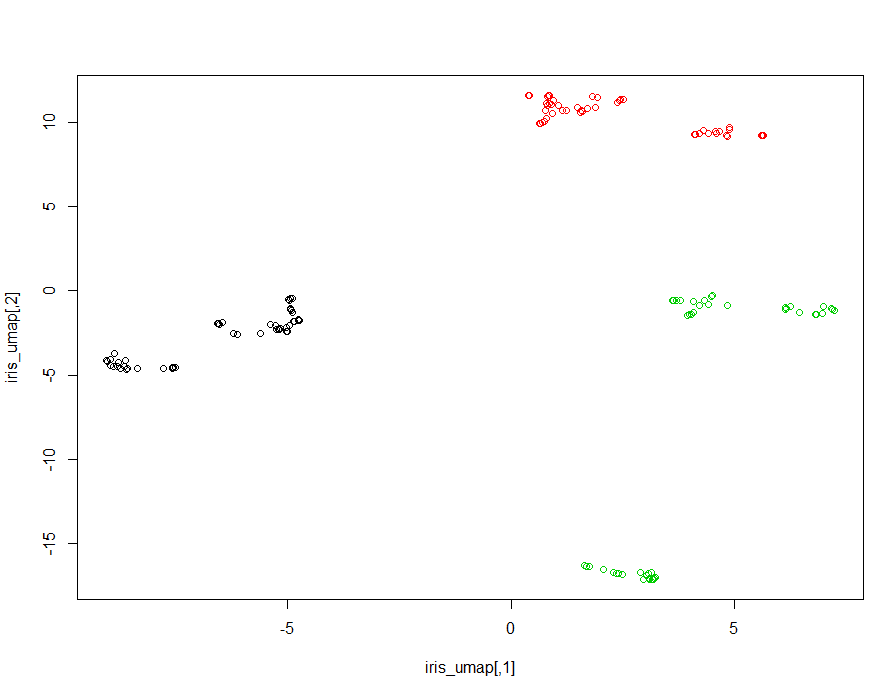
其降维分类效果比t-SNE更好,分的更开。
umap函数介绍
umap(X, n_neighbors = 15, n_components = 2, metric = "euclidean",
n_epochs = NULL, learning_rate = 1, scale = FALSE,
init = "spectral", init_sdev = NULL, spread = 1, min_dist = 0.01,
set_op_mix_ratio = 1, local_connectivity = 1, bandwidth = 1,
repulsion_strength = 1, negative_sample_rate = 5, a = NULL,
b = NULL, nn_method = NULL, n_trees = 50, search_k = 2 *
n_neighbors * n_trees, approx_pow = FALSE, y = NULL,
target_n_neighbors = n_neighbors, target_metric = "euclidean",
target_weight = 0.5, pca = NULL, pca_center = TRUE,
pcg_rand = TRUE, fast_sgd = FALSE, ret_model = FALSE,
ret_nn = FALSE, n_threads = max(1,
RcppParallel::defaultNumThreads()/2), n_sgd_threads = 0,
grain_size = 1, tmpdir = tempdir(), verbose = getOption("verbose",
TRUE))n_neighbors:确定相邻点的数量,通常其设置在2-100之间。
n_components:降维的维数大小,默认是2,其范围最好也在2-100之间。
Metric:距离的计算方法,有很多可以选择,具体的需要我们在应用的时候自行筛选。如:euclidean,manhattan,chebyshev,minkowski,canberra,braycurtis,mahalanobis,wminkowski,seuclidean,cosine,correlation,haversine,hamming,jaccard,dice,russelrao,kulsinski,rogerstanimoto,sokalmichener,sokalsneath,yule。
n_epochs:模型训练迭代次数。数据量大时200,小时500。
input:数据的类型,如果是data就会按照数据进行计算;如果dist就会认为是距离矩阵进行训练。
init:初始化用的。其中有这么三种方式: spectral,random,自定义。
min_dist:控制允许嵌入的紧密程度,值越小点越聚集,默认一般是0.1。
set_op_mix_ratio:设置降维过程中,各特征的结合方式,值0-1。0代表取交集,1代表取合集;中间就是比例。
local_connectivity:局部连接的点之间值,默认1,其值越大局部连接越多,导致的结果就是超越固有的流形维数出现改变。
bandwith:用于构造子集参数。
alpha:相当于在python中的leanging_rate(学习率)参数。
gamma:布局最优的学习率
negative_sample_rate:每一个阳性样本导致的阴性率。其值越大导致高的优化也就是过拟合,预测准确度下降。默认是5
spread:有效的嵌入式降维范围。与min_dist联合使用。
random_state:此值主要是确保模型的可重复性。如果不设置基于np.random,每次将会不同。
transform_seed:此值用于数值转换操作。一般默认42。
verbose: 控制工作日志,防止存储过多
参考资料:
1.https://cran.r-project.org/web/packages/umap/index.html
2.https://github.com/tkonopka/umap
3.https://arxiv.org/abs/1802.03426
4.https://github.com/lmcinnes/umap
5.https://blog.csdn.net/qq_36810544/article/details/81094469
6.https://github.com/jlmelville/uwot

 浙公网安备 33010802011761号
浙公网安备 33010802011761号
chenhao
更多详细细节,可以参考文末的文献和手册。