volcano3D可以探测三组之间差异表达的差异,其主要目的是为了在三维火山图中显示差异表达基因,这些图可以使用plotly转换为交互式可视化。其展现形式也让人耳目一新,下面简单介绍 volcano3D 的绘制。
为了方便演示,我们主要用到 volcano3D 、volcano3Ddata包:
install.packages("volcano3D")
devtools::install_github("KatrionaGoldmann/volcano3Ddata")1)构建 volcano3D 对象
library(volcano3D)
library(volcano3Ddata)
# 演示数据
data("syn_data")
# 构建volcano3D数据模型
syn_polar <- polar_coords(sampledata = syn_metadata, # 样本信息
# 信息选择,可以通过?polar_coords查看
contrast = "Pathotype",
pvalues = syn_pvalues,
expression = syn_rld,
p_col_suffix = "pvalue",
padj_col_suffix = "padj",
fc_col_suffix = "log2FoldChange",
multi_group_prefix = "LRT",
non_sig_name = "Not Significant",
significance_cutoff = 0.01,
label_column = NULL,
fc_cutoff = 0.1)2)绘制常规火山图
# 对volcano3D对象绘制火山图
syn_plots <-
volcano_trio(
polar = syn_polar,
sig_names = c("not significant","significant",
"not significant","significant"),
colours = rep(c("grey60", "slateblue1"), 2),
text_size = 9,
marker_size=1,
shared_legend_size = 0.9,
label_rows = c("SLAMF6", "PARP16", "ITM2C"),
fc_line = FALSE,
share_axes = FALSE)
3)绘制三维火山图俯视图
# 用plotly绘制可交互式图
radial_plotly(polar = syn_polar,
label_rows = c("SLAMF6", "PARP16", "ITM2C")) # 标记其中三个基因
# 用ggplot绘制静态图
radial_ggplot(polar = syn_polar,
label_rows = c("SLAMF6", "FMOD"),
marker_size = 2.3,
legend_size = 10)
4)同时也支持盒状图
boxplot_trio(syn_polar,
value = "FAM92B",
text_size = 7,
test = "polar_padj",
levels_order = c("Lymphoid", "Myeloid", "Fibroid"),
box_colours = c("blue", "red", "green3"),
step_increase = 0.1)
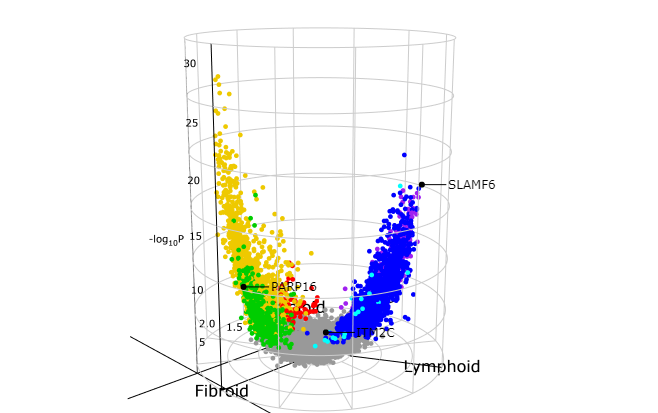
5)最后是3D火山图登场(单从意义上说没有什么大的亮点,但是从绘图的角度还是让人眼前一亮,后续三组比较的数据可以尝试)
# 参数此处不赘述了,可以查看下R文档,本函数的难点是在构建volcano3D对象上,
# 大家可以研究下,总体来说难度不大。
volcano3D(syn_polar,
label_rows = c("SLAMF6", "PARP16", "ITM2C"),
label_size = 10,
colour_code_labels = F,
label_colour = "black",
xy_aspectratio = 1,
z_aspectratio = 0.9,
plot_height = 600)
参考资料:
1.https://github.com/KatrionaGoldmann/volcano3D
2.https://peac.hpc.qmul.ac.uk/
 浙公网安备 33010802011761号
浙公网安备 33010802011761号